Welcome to MethFinder!
DNA methylation is a reversible epigenetic modification, and aberrant methylation disrupts gene regulation, driving tumorigenesis with cancer-type-specific patterns. Its reversibility and specificity make the identification of driver methylation events crucial for elucidating epigenetic mechanisms, discovering biomarkers, and revealing therapeutic targets. We introduce MethFinder, which can rapidly identify driver methylation at the site level. Additionally, this framework can be used to determine whether a target site is methylatable in cancer.
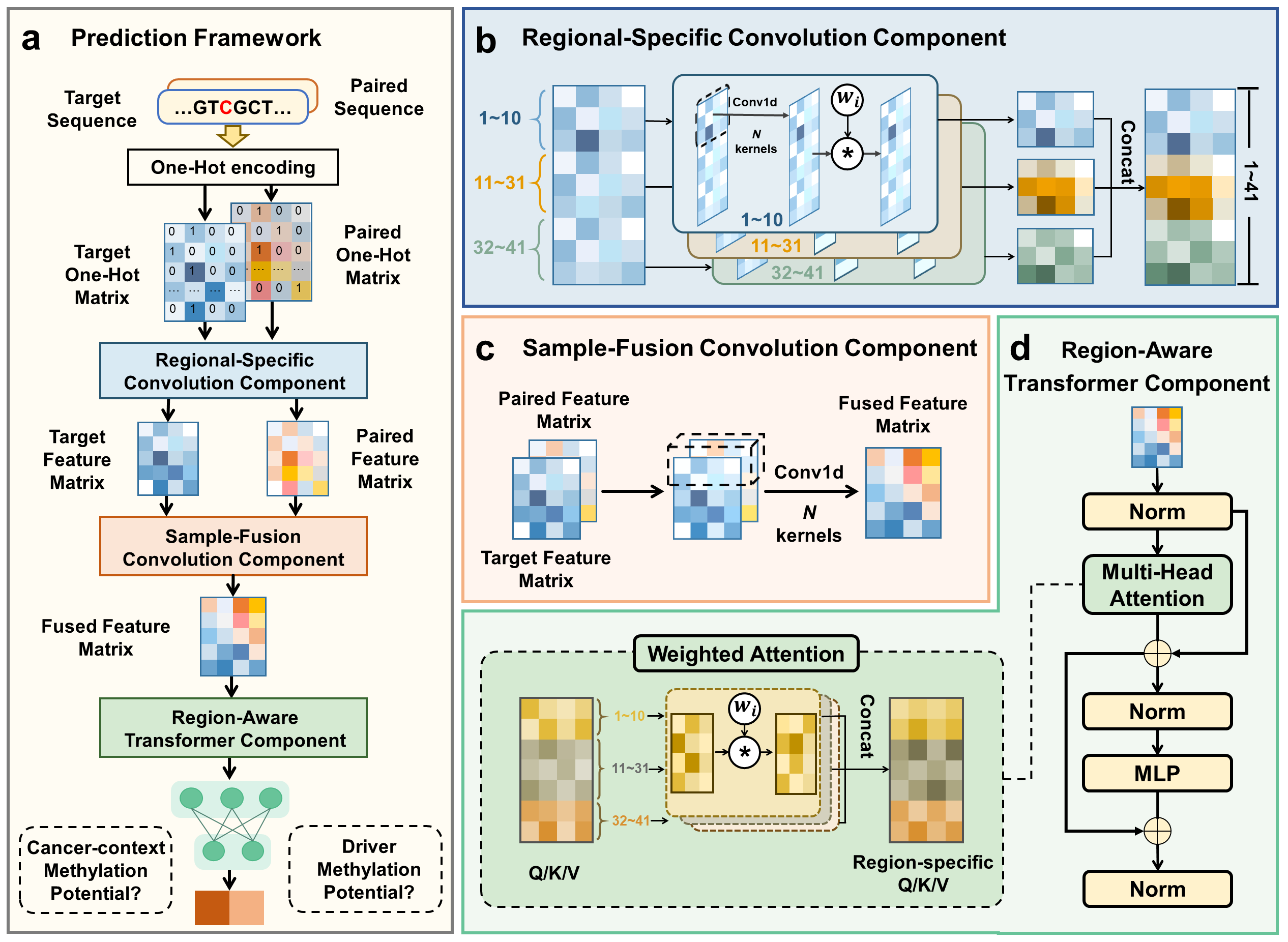
MethFinder consists of two modules, which respectively capture the potential of CpG sites to become methylated in the cancer context and their potential to drive specific cancer types. Both modules share a common architecture composed of three components (Fig. a). The model first converts each input sequence into a one-hot representation. It then begins with the RSC component, which divides the input sequence into upstream, core, and downstream regions and applies independent convolutional operations to each part to extract local regulatory features (Fig. b). The SFC component subsequently performs implicit contrastive learning by randomly introducing additional sample representations with the current one through convolutional fusion, thereby enhancing the model's robustness to data noises (Fig. c). Finally, the RAT component employs a Transformer encoder with region-weighted attention to model regulatory interactions among the local features and generate the final sequence representation (Fig. d). This representation is then fed into a classifier, enabling MethFinder to effectively learn the potential for CpG methylation in the cancer context and subsequently the potential of such methylation events to drive specific cancer types.
We provide researchers with a convenient and efficient MethFinder webserver for predicting driver methylations from sequence. Additionally, you can access the source code of MethFinder on our GitHub page https://github.com/yuyuan12138/MethFinder.
If you think MethFinder is useful, please kindly cite the following paper:
Webserver update:
Dec 26th, 2025: the first version of MethFinder server was established.
This work is
openly licensed via CC0 1.0