Welcome to MethFinder!
This online service provides three functions of MethFinder: including (1) predicting methylation sites that can occur in cancer contexts from any sequence, (2) identifying the type of cancer potentially driven by a given methylation site, and (3) predicting cancer type–specific driver methylation sites from any sequence.
The cancer types currently supported by MethFinder include:
| Abbreviation | Full Name |
|---|---|
| BLCA | Bladder Urothelial Carcinoma |
| BRCA | Breast Invasive Carcinoma |
| ESCA | Esophageal Carcinoma |
| HNSC | Head and Neck Squamous Cell Carcinoma |
| KIRC | Kidney Renal Clear Cell Carcinoma |
| KIRP | Kidney Renal Papillary Cell Carcinoma |
| LIHC | Liver Hepatocellular Carcinoma |
| LUAD | Lung Adenocarcinoma |
| LUSC | Lung Squamous Cell Carcinoma |
| PAAD | Pancreatic Adenocarcinoma |
| PCPG | Pheochromocytoma and Paraganglioma |
| PRAD | Prostate Adenocarcinoma |
| READ | Rectum Adenocarcinoma |
| SKCM | Skin Cutaneous Melanoma |
| THYM | Thymoma |
| UCEC | Uterine Corpus Endometrial Carcinoma |
We provide a user-friendly and robust webserver, enabling users to easily enable the above three functions based on MethFinder using DNA sequence in FASTA format. The simple usage tutorial is as follows:
-
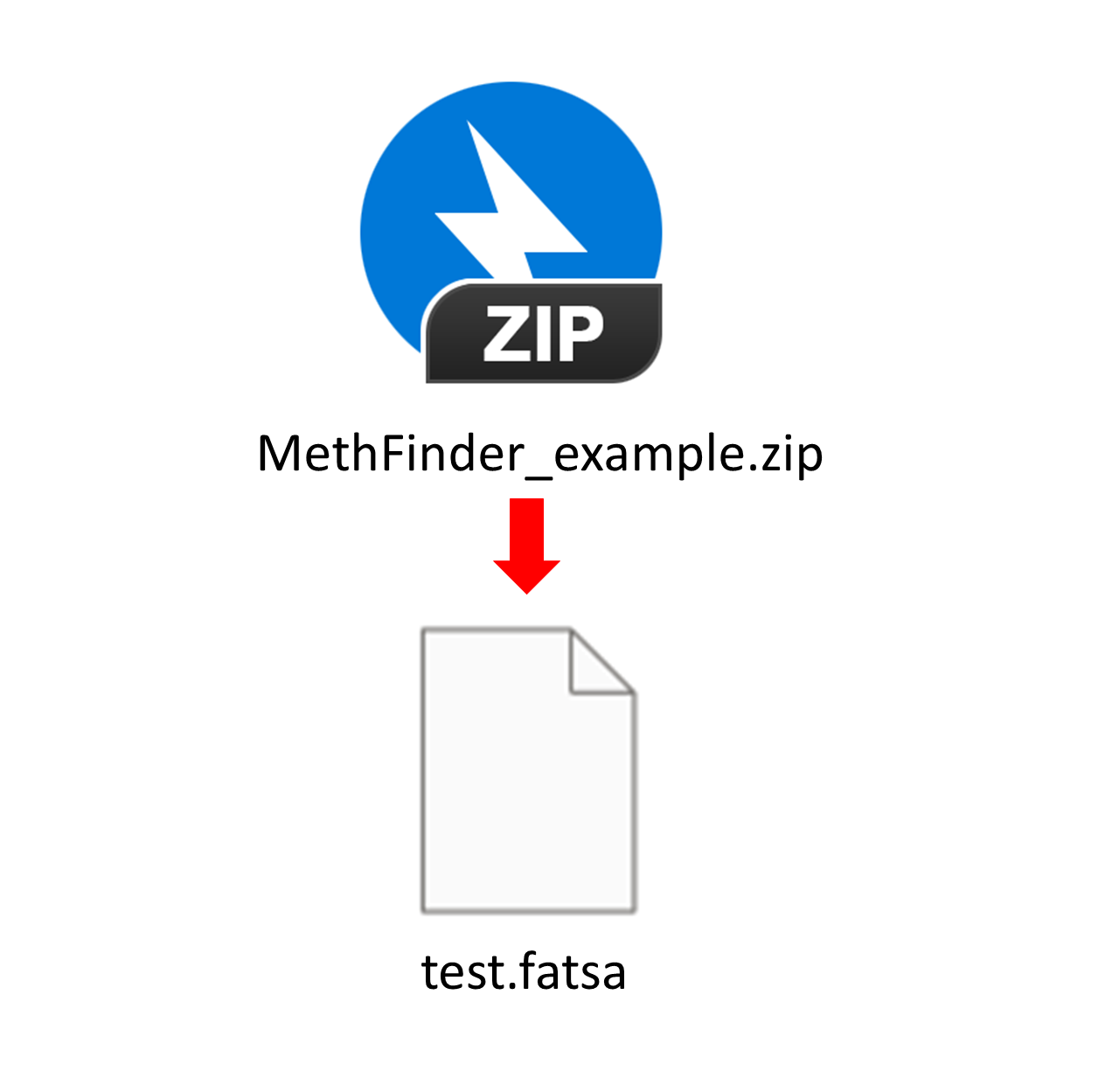
MethFinder Example data display:
The MethFinder online service accepts input files in .fasta format.
Each sequence must be 41 nucleotides long, and the central position
(i.e., the site to be predicted) must be a cytosine (C), as illustrated below.
After extracting the downloaded MethFinder_example.zip,
you will obtain an test.fasta file demonstrating the use of the online service.
-
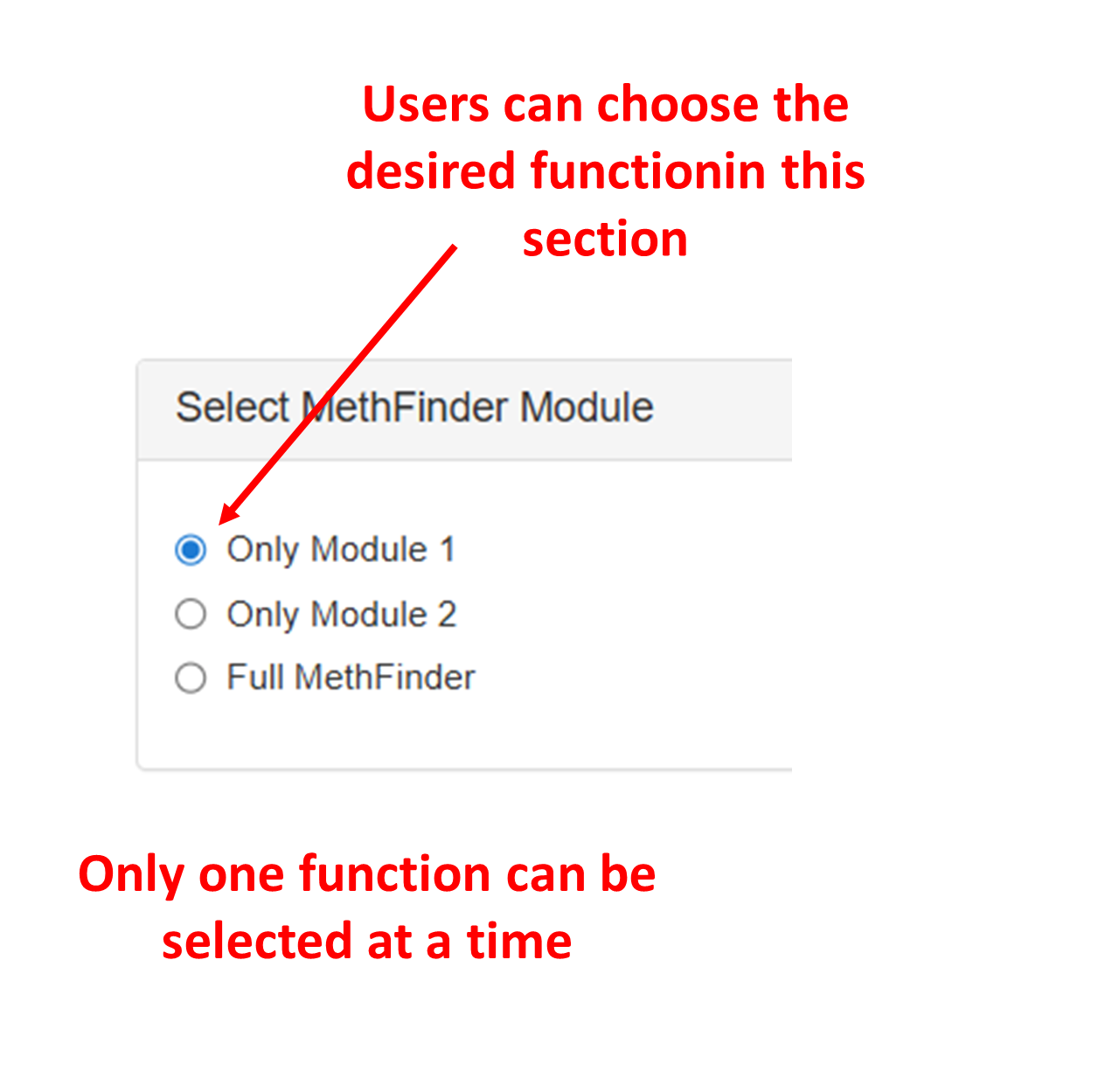
Choose the prediction mode:
-
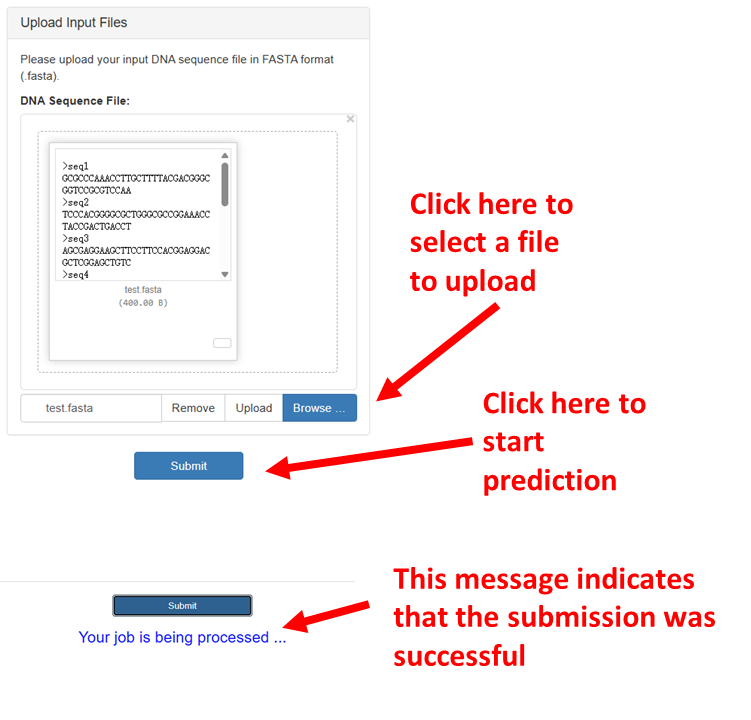
Upload the required FASTA files:
All uploaded files must be in FASTA format. MethFinder will automatically check for invalid input formats,
including sequence length and any illegal characters.
-
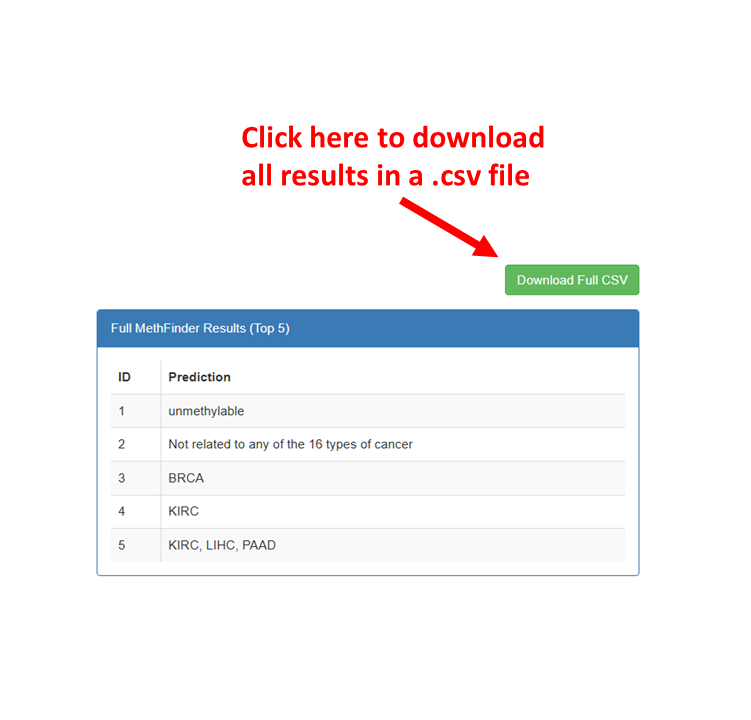
Interpretation of Results:
You will receive the results shown in the following figure.
"unmethylable" indicates that MethFinder predicts the site cannot be methylated in a cancer context.
"Not related ..." indicates that MethFinder predicts the site does not drive any cancer type
(limited to the 16 cancer types supported by MethFinder).
"KIRC, LIHC..." indicates the cancer type predicted by MethFinder to be driven by this site
(limited to the 16 cancer types supported by MethFinder).
-
MethFinder Example data display:
The MethFinder online service accepts input files in .fasta format. Each sequence must be 41 nucleotides long, and the central position (i.e., the site to be predicted) must be a cytosine (C), as illustrated below.
After extracting the downloaded MethFinder_example.zip, you will obtain an test.fasta file demonstrating the use of the online service.

-
Choose the prediction mode:

-
Upload the required FASTA files:
All uploaded files must be in FASTA format. MethFinder will automatically check for invalid input formats, including sequence length and any illegal characters.

-
Interpretation of Results:
You will receive the results shown in the following figure.
"unmethylable" indicates that MethFinder predicts the site cannot be methylated in a cancer context.
"Not related ..." indicates that MethFinder predicts the site does not drive any cancer type (limited to the 16 cancer types supported by MethFinder).
"KIRC, LIHC..." indicates the cancer type predicted by MethFinder to be driven by this site (limited to the 16 cancer types supported by MethFinder).

If you think MethFinder is useful, please kindly cite the following paper:
Webserver update:
This work is openly licensed via
CC0 1.0