NanoBind Server
NanoBind Server enables accurate prediction and comparison of nanobody–antigen interactions using advanced AI models.
NanoBind integrates five specialized sub-models to provide comprehensive analysis of nanobody–antigen interactions:
- NanoBind-seq: predicts nanobody–antigen interaction (NAI)
- NanoBind-pro: an enhanced version for refined NAI prediction
- NanoBind-site: predicts potential binding sites
- NanoBind-affi: estimates binding affinity levels
- NanoBind-pair: compares affinities between two nanobody–antigen pairs
Two operation modes are supported:
1. Standard Mode (NanoBind): integrates four models (seq + pro + site + affi) to predict NAI, binding sites, and affinities for a single nanobody–antigen pair.
2. Comparison Mode (NanoBind-pair): compares two nanobody–antigen pairs to determine which pair binds more strongly.
Detailed instructions and input requirements are available in the Readme file. For optimal performance, it is recommended not to upload more than 10 sequences simultaneously.
Source code:
https://github.com/zhaosq17/NanoBind
Datasets:
https://github.com/zhaosq17/NanoBind
Example files:
NanoBind_example.zip
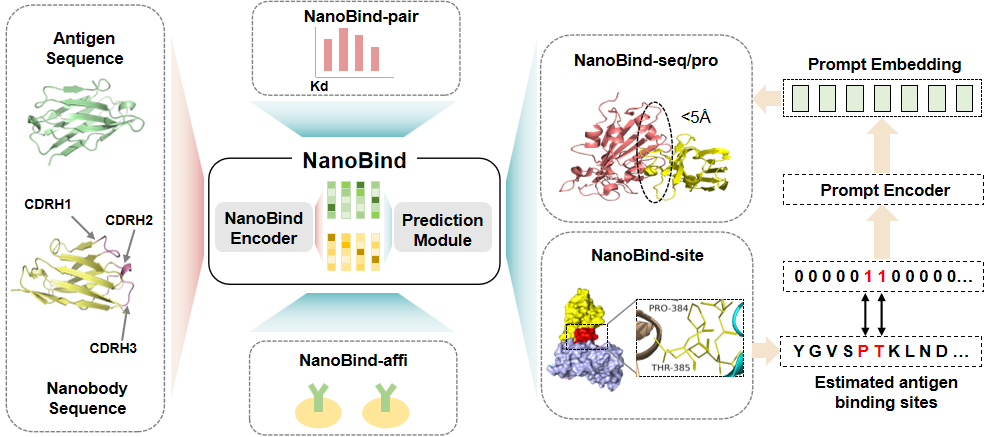
Figure: Overview of the NanoBind workflow.